Bio-Interfaces
Connecting a patterned substrate with biomolecules opens many new possiblities for assembling nanostructures. For example, snippets of DNA molecules can be used as "rubber bands" for
making connections between nanometer scale objects. DNA chips involve a patterned substrate which acts as a collection of miniature chemical beakers. Biosensors are being developed
that provide a field test for the presence of viruses,
using electrical or optical readout via liquid crystals or fluorescence.
Organic and biological molecules are typically attached to an inorganic surface in a multi-step process.
It starts with surface passivation layer, typically a self-assembled monolayer of alkanes with customized end groups.
Then comes a linker molecule which connects an end group to DNA or to an amino acid of a protein molecule.
By building up a such a scaffolding of multiple molecular layers it is possible to mimic the surface
of a living object.
To find out about the chemical bonds and the corresponding molecular orbitals in such layers we use
X-ray absorption spectroscopy, a technique based on
synchrotron radiation.
Below are a few examples of organic and biological molecules at surfaces, all the way to DNA and proteins.
For an introduction see: Xiaosong Liu et al.,
Can J. Chem. 85, 793 (2007).
Alkanethiols Adsorbed on Gold-Coated Silicon
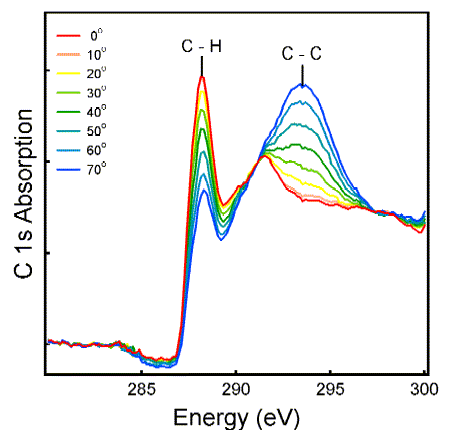
Alkane chains passivate the surface and prevent biomolecules from denaturing. The orientation of the chains is obtained by polarized synchrotron radiation using dipole selection rules. These are rather simple for transitions from the C 1s core level to the C 2p valence orbitals: The intensity is highest when the
polarization points parallel to the orbital, and it follows a cos2(theta) pattern. The polarization angle theta relative to the sample surface is indicated in the spectra below.
The C-H orbitals lie nearly parallel to the surface, whereas the C-C orbitals of the backbone are nearly perpendicular. That implies a perpendicular orientation of the molecules.
A more quantitative analysis shows that there is a range of tilt angles around this average orientation.
Self-Assembled Monolayers of Siloxanes
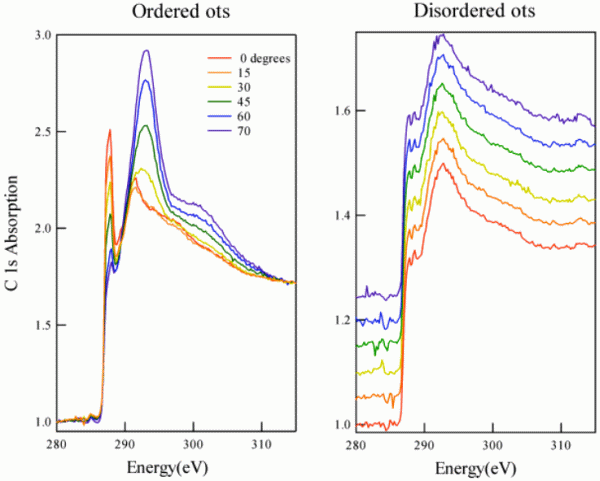
Attaching alkane chains via siloxane chemistry is more direct, but also more delicate. The role of humidity and settling time in preparing a well-ordered self-assembled monolayer (SAM)
is demonstrated here for octadecyltrichlorosilane (OTS).
The set of polarization-dependent X-ray absorption spectra on the left is from a SAM that had enough humidity and time to order itself, while the spectra on the right are from a quickly-dried layer. The polarization dependence has disappeared on the right.
(The spectra are offset from each other, because otherwise they would collapse to the same curve.) The lack of order is connected with a reduced coverage, as reflected by the reduced absorption scale on the left. This observation is consistent with a model where crowded molecules form an ordered SAM to increase their packing density.
R. Peters et al.,
Langmuir 18, 1250 (2002).
Oriented DNA Snippets at a Surface
The same type of spectroscopy can be applied to more complex molecules, such as DNA segments. The nitrogen 1s core level is chosen in order to focus onto the nucleotide bases, the only part of DNA containing nitrogen. The base pairs contain characteristic pi* orbitals oriented perpendicular to the pi-rings. That makes them parallel to the axis of the double-helix. From the polarization dependence, one can tell that
the DNA molecules are partially oriented and that the axis of the double-helix lies perpendicular to the surface (on average).
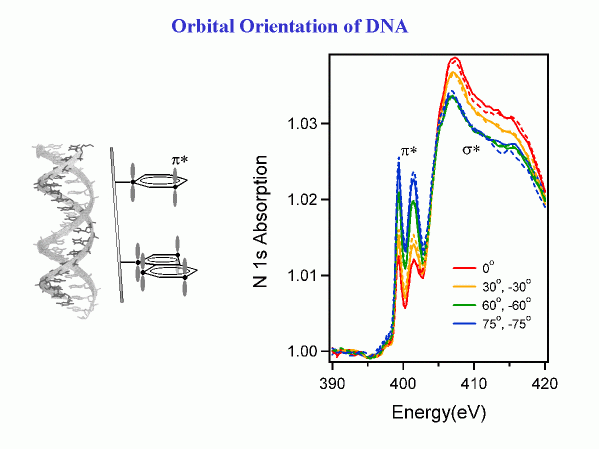
Systematic work with single-stranded DNA shows how the degree of orientation depends on the length of the DNA molecules and on the bases (collaboration with NRL and NIST). The orientation of DNA at a surface is important for designing DNA microarrays.
A single-stranded DNA molecule attached to a surface needs to stick out to allow a complementary strand to bind to it.
D. Y. Petrovykh et al.,
J. Am. Chem. Soc. 128, 2 (2006) , supporting information.
Oriented Proteins at a Surface
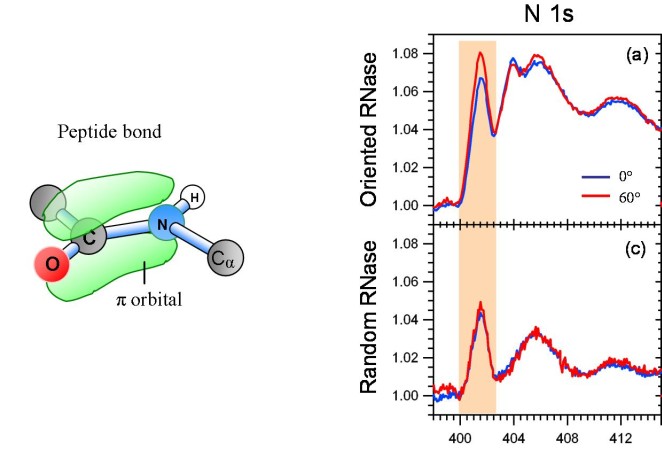
Going farther up in complexity we have tackled small proteins, such as RNase A. It consists of 124 amino acids (about a thousand atoms not counting H).
In X-ray absorption spectroscopy, the characteristic orbital of proteins is the pi* orbital of the peptide bond, which connects the amino acids to form the
backbone of the protein. As with DNA, the average orientation of the peptide bonds can be determined from the polarization dependence of the
X-ray absorption at the N 1s edge. A modulation of 20% was found for this orbital when RNase A was immobilized in oriented fashion. That is
consistent with a calculation based on the crystallographic data in the protein data base.
When the protein is immobilized in random orientation, the polarization dependence vanishes. Oriented immobilization of proteins is essential for
the functioning of biosensors, because the docking site has to point outwards in order to link to a target virus.
Xiaosong Liu et al.,
Langmuir 22, 7719 (2006).
Universal Mechanism for Breaking Peptide Bonds
The peptide bond (an amide bond) plays a central role in protein chemistry. It forms the backbone of proteins by linking amino acids and thus determines the chemical stability of all forms of life. The effect of radiation on biomolecules is of great interest for many reasons, among them radiation-induced mutations, UV-induced skin cancer, radiation therapy, irradiation of food for sterilization, radiation damage by x-rays in protein crystallography, proteomics via electrospray ionization, radiation damage of biological samples in microscopy, and the role of radiation in forming the chemical building blocks of life.
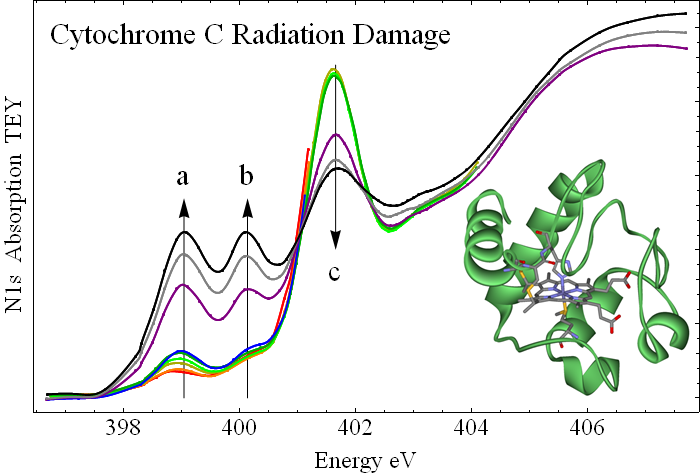
X-ray absorption spectroscopy reveals a universal photochemical bond breaking pattern of amides, which leaves a characteristic fingerprint at the N 1s X-ray absorption edge in the form of two low-lying pi* peaks (a and b in the figure). These are identified with imine and nitrile groups by searching through many model compounds. Pristine cytochrome has a single pi* peak (c in the figure) which is characteristic of the peptide bond orbital (see the previous figure). This peak shrinks with irradiation, showing that peptide bonds are broken.
P. S. Johnson et al.,
J. Chem. Phys. 135, 044702 (2011).
P. L. Cook et al.,
J. Chem. Phys. 131, 214702 (2009).
Supported by
NSF-DMR,
NSF-CHE, and
DOE-BES.
Franz Himpsel's Home Page




